4 Post-Process Image Segmentation
Below is a walk-through for the post-process image segmentation (i.e., blob identification) of iLAM images to identify movements
1. Organize your file directory structure in a logical, hierarchical structure
~/iLAM/exp_a/contains images, metadata, and analysis scripts specific to Experiment A- Experiment A’s images from ilam_01 and ilam_02 are saved in
~/iLAM/exp_a/ilam_01/and~/iLAM/exp_a/ilam_02/, respectively - .CSV output containing the size, location, and timing of all blobs/movements for each iLAM cages for Experiment A (i.e., ilam_01 and ilam_02):
~/iLAM/exp_a/iLAM_exp_a.txt/, with corresponding metadata:~/iLAM/exp_a/metadata_exp_a.csv/
## Warning: package 'data.tree' was built under R version 4.3.3## levelName
## 1 iLAM
## 2 ¦--exp_a
## 3 ¦ ¦--ilam01
## 4 ¦ ¦ ¦--ilam01.0707.2058.jpg
## 5 ¦ ¦ °--ilam01.0707.2100.jpg
## 6 ¦ ¦--ilam02
## 7 ¦ ¦ ¦--ilam02.0707.2058.jpg
## 8 ¦ ¦ °--ilam02.0707.2100.jpg
## 9 ¦ ¦--scripts
## 10 ¦ ¦ ¦--analyze_experiment.R
## 11 ¦ ¦ ¦--exp_a_ilam01.R
## 12 ¦ ¦ ¦--exp_a_ilam02.R
## 13 ¦ ¦ ¦--run_exp_a_ilam01.sh
## 14 ¦ ¦ °--run_exp_a_ilam02.sh
## 15 ¦ ¦--metadata_exp_a.csv
## 16 ¦ °--iLAM_exp_a.txt
## 17 ¦--exp_b
## 18 ¦ ¦--ilam01
## 19 ¦ ¦--ilam02
## 20 ¦ °--scripts
## 21 °--scripts
## 22 ¦--add_columns_to_42.R
## 23 ¦--find_movements.R
## 24 ¦--find_threshold.R
## 25 ¦--make_dam_file.R
## 26 ¦--make_gif.R
## 27 ¦--parse_movements.R
## 28 °--plot_movements.R2. Install iLAMtools from GitHub
Alternatively, you can download iLAM functions from this zip folder: iLAM wrapper functions and reference them with ‘source()’.
To verify installation, a zip folder containing a set of sample images and test script can be downloaded here (198MB): iLAM_test folder
2. Perform image segmentation to identify movements across images taken by each iLAM (exp_a_ilam01.R, exp_a_ilam02.R, etc.)
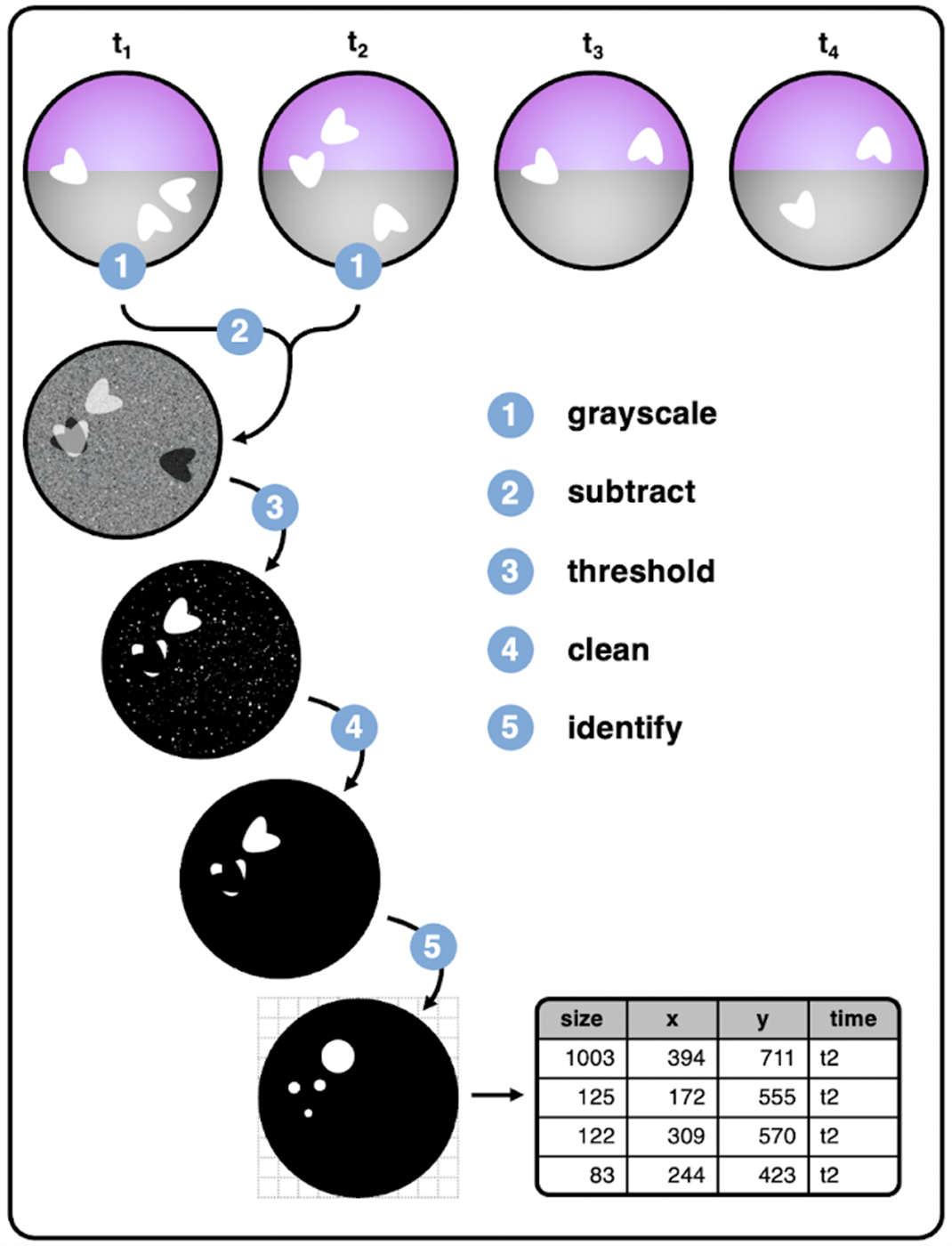
Figure 4.1: Image segmentation workflow
- Load required packages and iLAM functions
setwd("/~/iLAM/exp_a/")
library(iLAMtools)- Update and tailor values for every cage and/or experiment; these values are recorded to metadata output and used as input settings for the find_movements() iLAM wrapper function:
- Cage-Specific: Integer coordinates for image cropping (x_left, x_right, y_bot, y_top)
- Experiment-Specific:
- n_thr: Numeric threshold percentile; higher values indicate a more stringent filtering. e.g., at n_thr=0.999, only the darkest pixel differences >99.9% are retained for blob identification (default: n_thr = 0.996)
- Note: For moths (O. nubilalis, H. zea, S. frugiperda), we use: n_thr=0.999
- Note: For beetles (Photinus species, H. axyridus), we use: n_thr=0.999
- n_cln: Integer value to clean up pixels: e.g., at n_cln = 5, this first “shrinks” to remove all isolated blobs smaller than 5 pixels, and then “grows” remaining blobs by 5*n_grw pixels to aggregate nearby blobs with each other. (default: n_cln = 5, n_grw = 1.5)
- Note: For moths (O. nubilalis, H. zea, S. frugiperda), we use: n_cln=10, n_grw=1.5
- Note: For beetles (Photinus species, H. axyridus), we use: n_cln=10, n_grw=1.5
- genus: Character string for study organism
- species: Character string for study organism
- color: Character string to indicate whether the study organism appears “white” or “black” on the background
- Note: For moths (O. nubilalis, H. zea, S. frugiperda), we use: “white”
- Note: For beetles (Photinus species), we use: “black”
- For more information on threshold, clean, blur, etc., see imager documentation for more details.
- n_thr: Numeric threshold percentile; higher values indicate a more stringent filtering. e.g., at n_thr=0.999, only the darkest pixel differences >99.9% are retained for blob identification (default: n_thr = 0.996)
pi_sub_folder <- "ilam01"
sex <- "male" #cage/project-specific
#Crop-settings
x_left <- 425 #cage-specific, depending on camera arrangement
x_right <- 2225 #cage-specific, depending on camera arrangement
y_bot <- 100 #cage-specific, depending on camera arrangement
y_top <- 1700 #cage-specific, depending on camera arrangement
#change following values for every experiment
out_file_name = "iLAM_exp_a" #project-specific
n_thr = 0.999 #species-specific, depending on IR reflectance/contrast with background
n_cln = 10 #species-specific, depending on IR reflectance
n_max = 75000 #species-specific, pixel differences above this value will be considered as noise
start_photophase = 5 #project-specific, time that lights turn on
end_photophase = 21 #project-specific, time that dark starts
genus = "photinus" #project-specific
species = "marginellus" #project-specific
animal = "black" #project-specific, during the night, does the animal appear "white" on a dark background or "black" on a light background? This is VERY important (!) because it determines whether "movements" identify insects whom left vs. arrived between frames - Create a vector of .jpg image file names to be analyzed
file_names <- list.files(pi_sub_folder,
pattern= "*.jpg",
full.names = TRUE)We identify cropping locations that remove the outer edges of the cage, and maintain a constant picture area across all iLAMs (e.g., 1800x1600 pixels) with the following command:
load.image(file_names[1]) %>%
imsub(x %inr% c(x_left,x_right), y %inr% c(y_bot,y_top)) %>% plot()Find all movements by image subtraction, global thresholding, and blob detection in the iLAM wrapper function
find_movements().-Additional optional settings:
- n_blr: Integer value to set blur radius to denoise image and reduce graininess, prior to image subtraction (default: n_blr = 3)
- n_grw: Integer value for n_cln multiplier to “grow” and aggregate blobs within a given proximity. For example, when n_cln=5 and n_grw = 1.5, blobs are first shrunk by 5 pixels and then grown by 5x1.5=7.5 pixels (default: n_grw = 1.5)
- n_max: Integer value denoting the maximum number of pixel differences expected between two images; any values above this n_max will be considered as noisy and assigned an arbitrary value for subsequent filtering/removal. See @ref(04-make_dam_output.Rmd) for more information. Example, if a moth movement = 3000 pixels and there are five moths in a cage, then the maximum number of differences expected is 15,000 pixels. (default: n_max = 75000)
- find_thr: Boolean value “T” (true) or “F” (false) to determine what pixel value cut-off corresponds to the n_thr percentile difference across a p_sample of image comparisons (default = “T”)
- type_thr: String value “absolute” or “relative” indicating whether to perform absolute (capture all differences > n_thr for all pictures) or relative thresholding procedure (capture all differences > n_thr for each comparison) on set of images. We recommend the default, “absolute” option because the latter identifies false movements in images where there are no movements because it always retains the top pixel differences. (default: find_thr = “absolute”)
- p_sample: Numeric value denoting the proportion of image comparisons that should be used to determine the absolute threshold value
- Note: If 5% of the total images is <100, then increasing p_sample >0.05 is recommended
- channel: String value denoting which color channel movements should be identified from (default: “grayscale”)
out <- find_movements(files = file_names, # list of file names
n_thr = n_thr, # threshold value (0.992 == "0.8%")
n_cln = n_cln, # value for cleaning (number of pixels)
n_grw = 1.5, # multiplier for n_cln (shrink vs. grow)
n_blr = 3, # let user select blur radius
n_max = 75000, # upper cut-off for # pixel differences
x_left = x_left, # value for crop on x min
x_right = x_right, # value for crop on x max
y_bot = y_bot, # value for crop on y min
y_top = y_top, # value for crop on y max
find_thr = T, # T or F
type_thr = "absolute",
p_sample = 0.05, # If 5% of the total images < 100, then increase this value
channel = "grayscale",
animal = animal)- Update and save output containing all identified blobs, their size and location as a .csv in the current working directory.
#adds additional columns to dataframe
out$ID <- paste0(n_thr*100,"%_", "s", n_cln, "g", 1.5*n_cln)
out$sex <- sex
out$genus <- genus
out$species <- species
if (file.exists(paste0(out_file_name,".csv"))){
write.table(out, file = paste0(out_file_name,".csv"),
append = TRUE, quote = TRUE, sep = ",",
row.names = FALSE, col.names = FALSE)
} else{
write.csv(out, file = paste0(out_file_name,".csv"),
col.names = TRUE, row.names = FALSE)
}
rm(out)- Use
plot_movements()to visualize movements by plotting detected blobs onto a subset of images. These images are helpful for tuning parameters.
#circles in bottom left corner denote standards of sizes: 12800 px, 3200 px, 800 px, 200 px, 50 px
plot_movements(file_names,
pi_sub_folder,
by_change,
x_left, x_right,
y_bot, y_top,
n_max)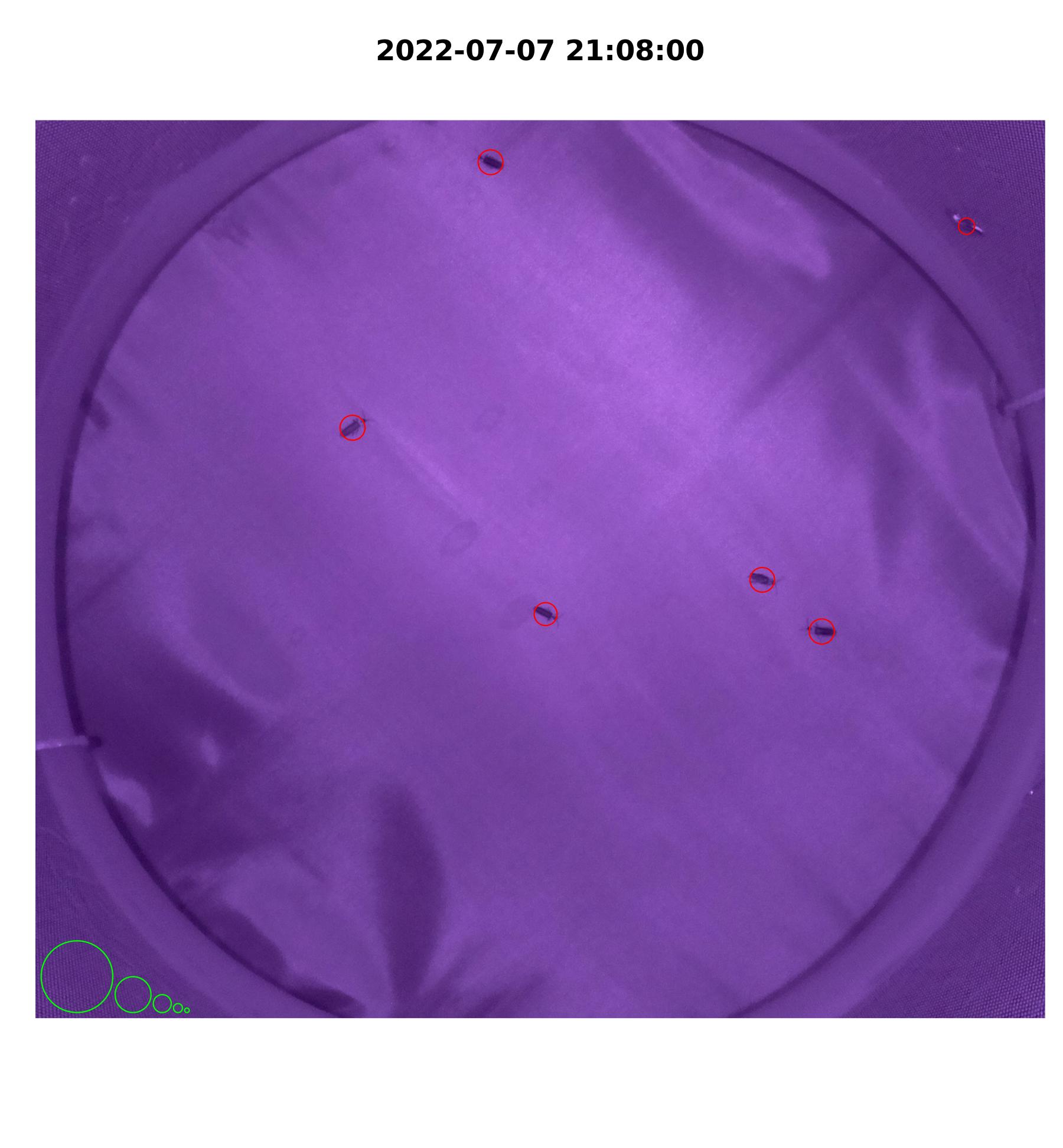
Figure 4.2: iLAM plot_movements() image
- If desired, make a gif from plotted images with
make_gif()
library(magick)
make_gif(out_file_name,
pi_sub_folder)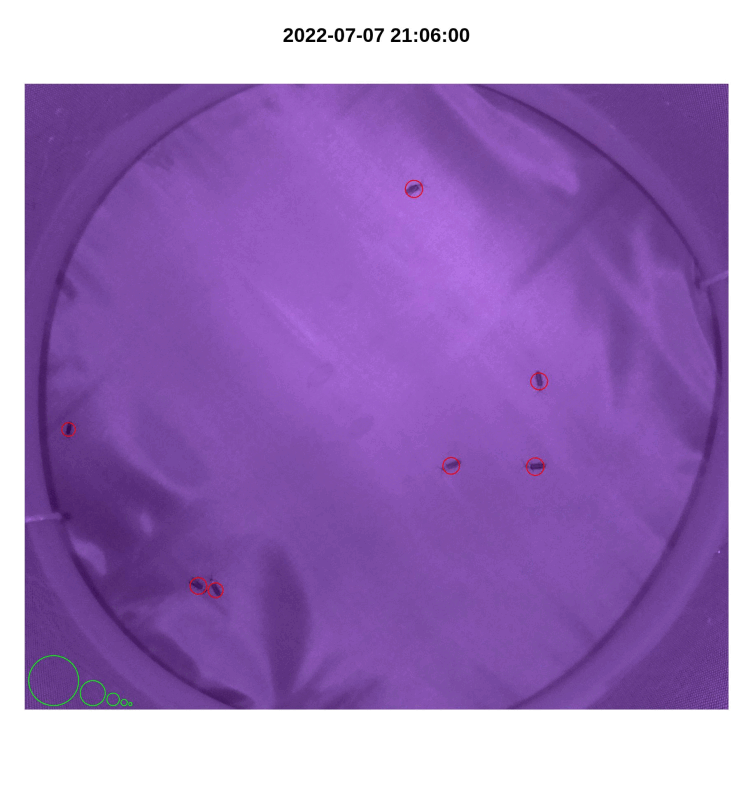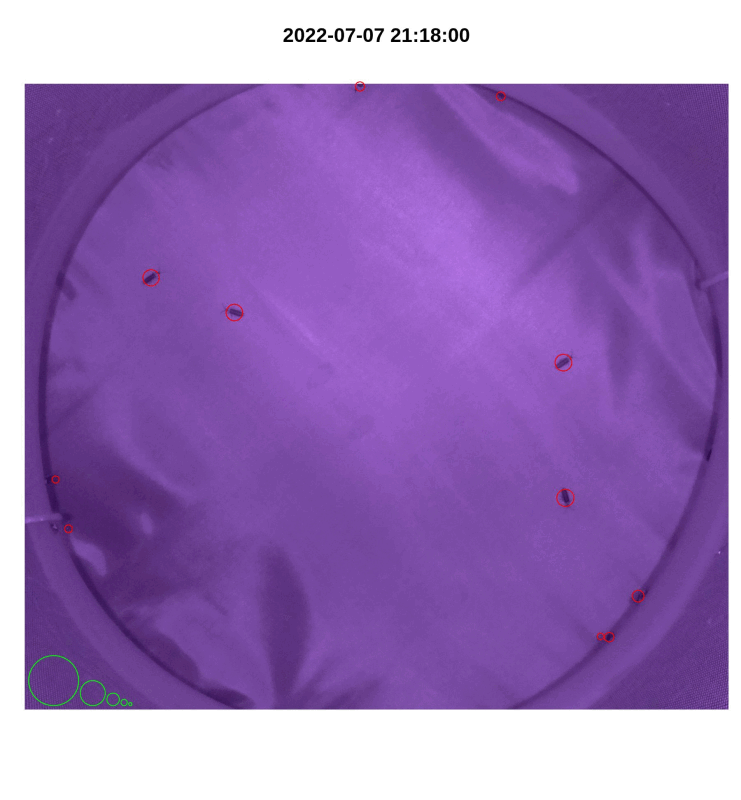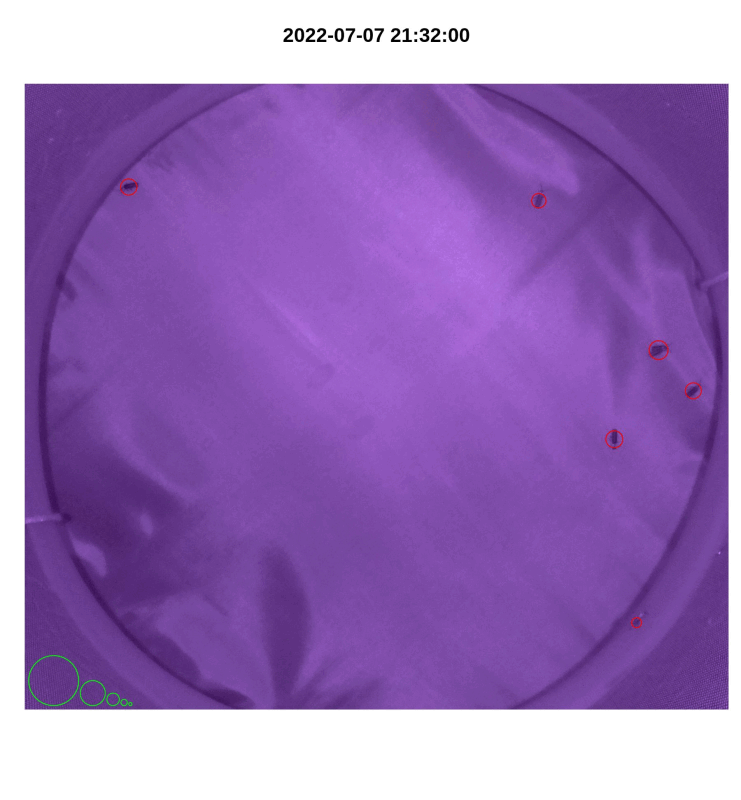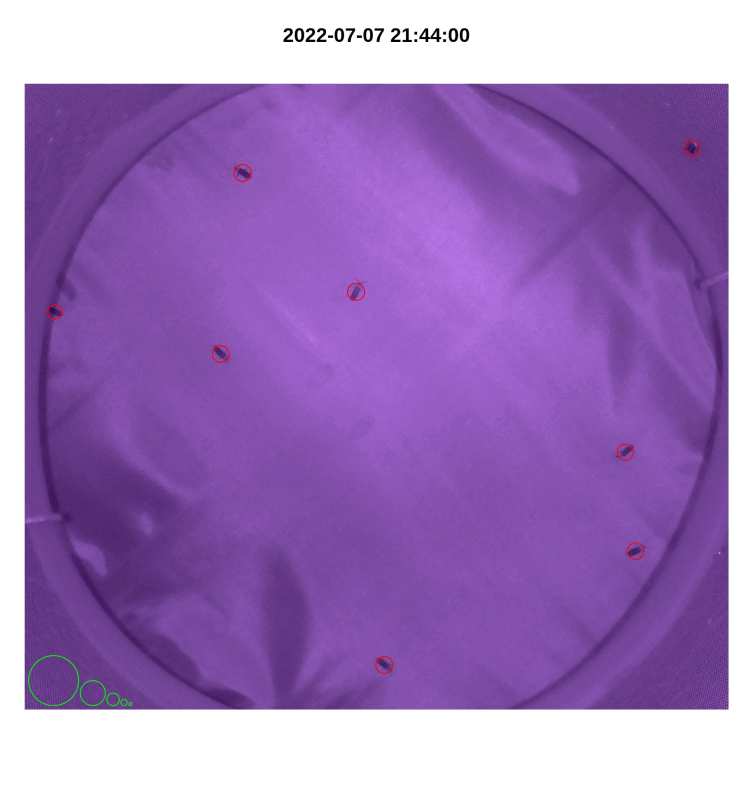
Figure 4.3: iLAM make_gif() output
Tips:
- First test parameters settings for
find_movements()with a subset of experimental images: e.g.,find_movements(files = file_names[1:1000], p_sample=0.10) - If
find_movements()crashes, returns excess small blobs/movements, and/or returns excess blobs/movements that do NOT correspond to REAL movements, then increase the n_thr or p_sample values. If this issue continues, then remove any days at the experiment end where all/most insects have died; this can downwardly bias the average number of pixel differences expected across images. - If
find_movements()returns many small blobs/animal and you desire one blob/animal movement, then increase n_cln and/or n_grw